Last month, I announced that we would soon be making EC2 Dedicated Hosts available. As I wrote at the time, this model allows you to control the mapping of EC2 instances to the underlying physical servers. Dedicated Hosts allow you to:
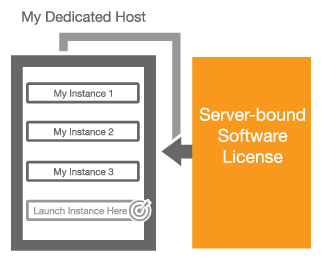 Bring Your Own Licenses – You can bring your existing server-based licenses for Windows Server, SQL Server, SUSE Linux Enterprise Server, and other enterprise systems and products to the cloud. Dedicated Hosts provide you with visibility into the number of sockets and physical cores that are available so that you can obtain and use software licenses that are a good match for the actual hardware.
Bring Your Own Licenses – You can bring your existing server-based licenses for Windows Server, SQL Server, SUSE Linux Enterprise Server, and other enterprise systems and products to the cloud. Dedicated Hosts provide you with visibility into the number of sockets and physical cores that are available so that you can obtain and use software licenses that are a good match for the actual hardware.- Help Meet Compliance and Regulatory Requirements – You can allocate Dedicated Hosts and use them to run applications on hardware that is fully dedicated to your use.
- Track Usage – You can use AWS Config to track the history of instances that are started and stopped on each of your Dedicated Hosts. This data can be used to verify usage against your licensing metrics.
- Control Instance Placement – You can exercise fine-grained control over the placement of EC2 instances on each of your Dedicated Hosts.
Available Now
I am happy to be able to announced the Dedicated Hosts are available now and that you can start using them today. You can launch them from the AWS Management Console, AWS Command Line Interface (CLI), AWS Tools for Windows PowerShell, or via code that makes calls to the AWS SDKs.
Let’s provision a Dedicated Host and then launch some EC2 instances on it via the Console! I simply open up the EC2 Console, select Dedicated Hosts in the left-side navigation bar, and click on Allocate a Host.
I choose the instance type (Dedicated hosts for M3, M4, C3, C4, G2, R3, D2, and I2 instances are available), the Availability Zone, and the quantity (each Dedicated Host can accommodate one or more instances of a particular type, all of which must be the same size).
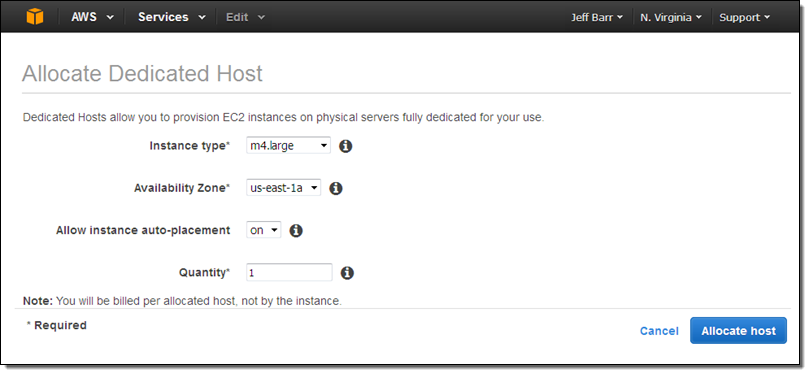
If I choose to allow instance auto-placement, subsequent launches of the designed instance type in the chosen Availability Zone are eligible for automatic placement on the Dedicated Host, and will be placed there if instance capacity is available on the host and the launch specifies a tenancy of Host without specifying a particular one. If I do not allow auto-placement, I must specifically target this Dedicated Host when I launch an instance.
When I click Allocate host, I’ll receive confirmation that it was allocated:
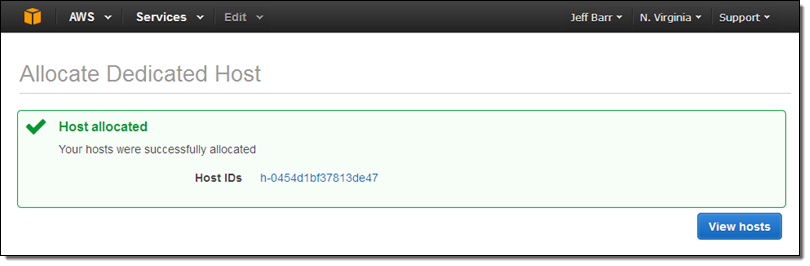
Billing for the Dedicated Host begins at this point. The size and number of instances are running on it does not have an impact on the cost.
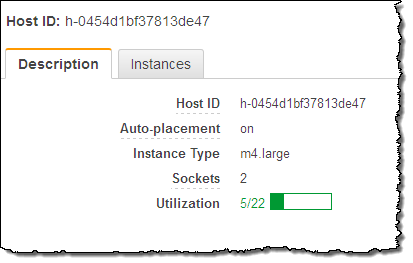
I can see all of my Dedicated Hosts at a glance. Selecting one displays detailed information about it:
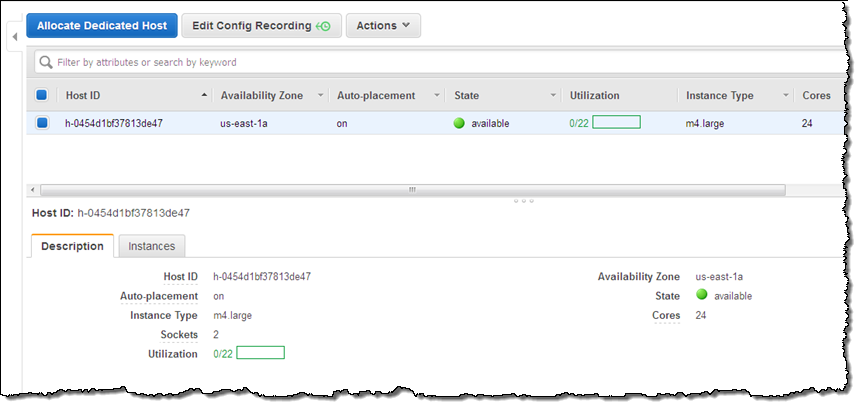
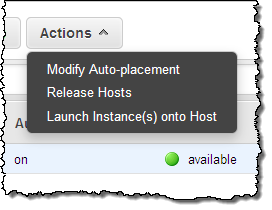
As you can see, my Dedicated Host has 2 sockets and 24 cores. It can host up to 22 m4.large instances, but is currently not hosting any. The next step is run some instances on my Dedicated Host. I click on Actions and choose Launch Instance(s) onto Host (I can also use the existing EC2 launch wizard):
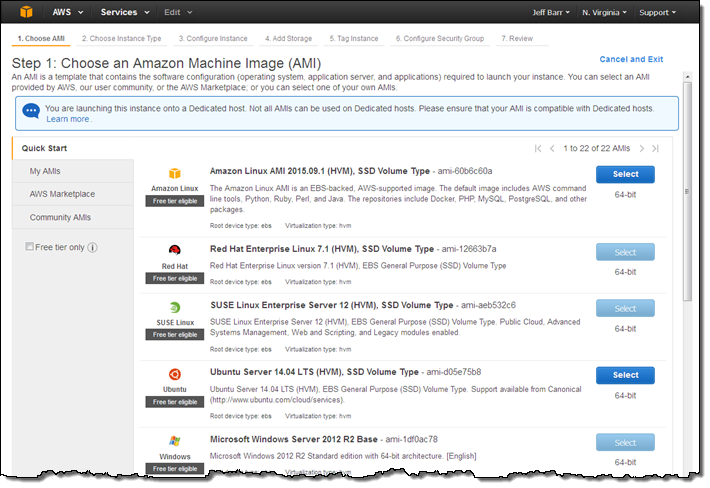
Then I pick an AMI. Some AMIs (currently RHEL, SUSE Linux, and those which include Windows licenses) cannot be used with Dedicated Hosts, and cannot be selected in the screen below or from the AWS Marketplace:
The instance type is already selected:
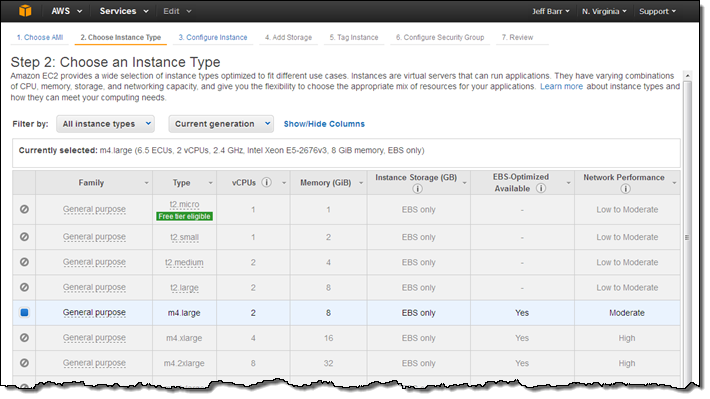
Instances launched on a Dedicated Host must always reside within a VPC. A single Dedicated Host can accommodate instances that run in more than one VPC.
The remainder of the instance launch process proceeds in the usual way and I have access to the options that make sense when running on a Dedicated Host. You cannot, for example, run Spot instances on a Dedicated Host.
I can also choose to target one of my Dedicated Hosts when I launch an EC2 instance in the traditional way. I simply set the Tenancy option to Dedicated host and choose one of my Dedicated Hosts (I can also leave it set to No preference and have AWS make the choice for me):
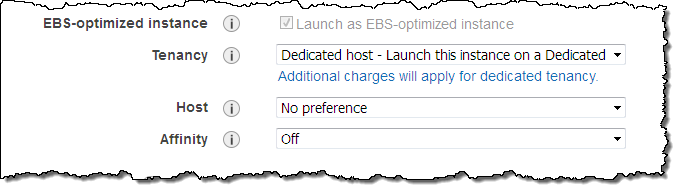
If I select Affinity, a persistent relationship will be created between the Dedicated Host and the instance. This gives you confidence that the instance will restart on the same Host, and minimizes the possibility that you will inadvertently run licensed software on the wrong Host. If you import a Windows Server image (to pick one that we expect to be popular), you can keep it assigned to a particular physical server for at least 90 days, in accordance with the terms of the license.
I can return to the Dedicated Hosts section of the Console, select one of my Hosts, and learn more about the instances that are running on it:
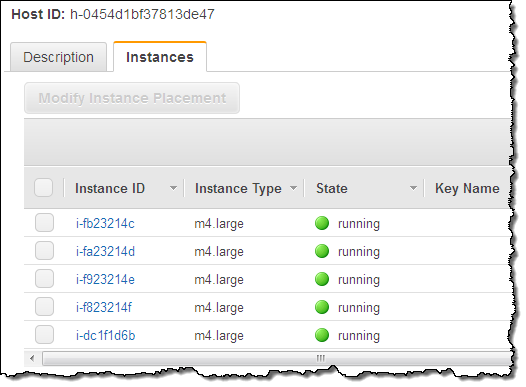
Using & Tracking Licensed Software
You can use your existing software licenses on Dedicated Hosts. Verify that the terms allow the software to be used in a virtualized environment, and use VM Import/Export to bring your existing machine images into the cloud. To learn more, read about Bring Your Own License in the EC2 Documentation. To learn more about Windows licensing options as they relate to AWS, read about Microsoft Licensing on AWS and our detailed Windows BYOL Licensing FAQ.
You can use AWS Config to record configuration changes for your Dedicated Hosts and the instances that are launched, stopped, or terminated on them. This information will prove useful for license reporting. You can use the Edit Config Recording button in the Console to change the settings (hovering your mouse over the button will display the current status):
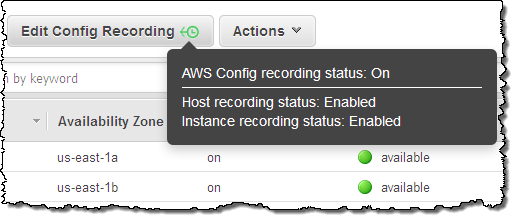
To learn more, read about Using AWS Config.
Some Important Details
As I mentioned earlier, billing begins when you allocate a Dedicated Host. For more information about pricing, visit the Dedicated Host Pricing page.
EC2 automatically monitors the health of each of your Dedicated Hosts and communicates it to you via the Console. The state is normally available; it switches to under-assessment if we are exploring a possible issue with the Dedicated Host.
Instances launched on Dedicated Hosts must always reside within a VPC, but cannot make use of Placement Groups. Auto Scaling is not supported, and neither is RDS.
Dedicated Hosts are available in the US East (Northern Virginia), US West (Oregon), US West (Northern California), Europe (Ireland), Europe (Frankfurt), Asia Pacific (Tokyo), Asia Pacific (Singapore), Asia Pacific (Sydney), and South America (Brazil) regions. You can allocate up to 2 Dedicated Hosts per instance family (M4, C4, and so forth) per region; if you need more, just ask.
— Jeff;


 Bring Your Own Licenses – You can bring your existing server-based licenses for Windows Server, SQL Server, SUSE Linux Enterprise Server, and other enterprise systems and products to the cloud. Dedicated Hosts provide you with visibility into the number of sockets and physical cores that are available so that you can obtain and use software licenses that are a good match for the actual hardware.
Bring Your Own Licenses – You can bring your existing server-based licenses for Windows Server, SQL Server, SUSE Linux Enterprise Server, and other enterprise systems and products to the cloud. Dedicated Hosts provide you with visibility into the number of sockets and physical cores that are available so that you can obtain and use software licenses that are a good match for the actual hardware.









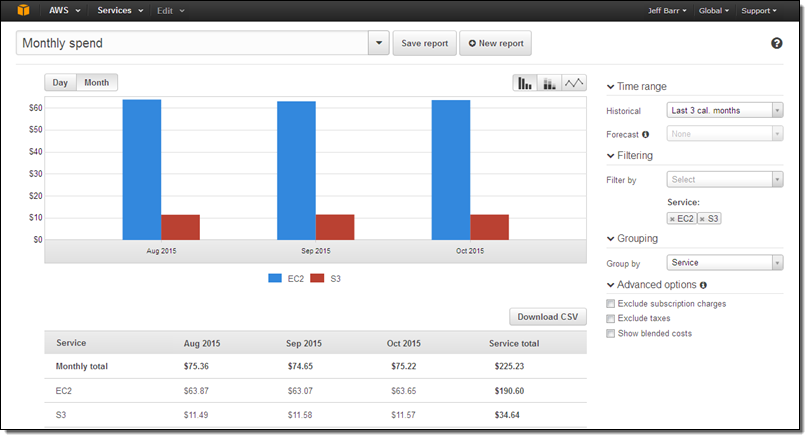
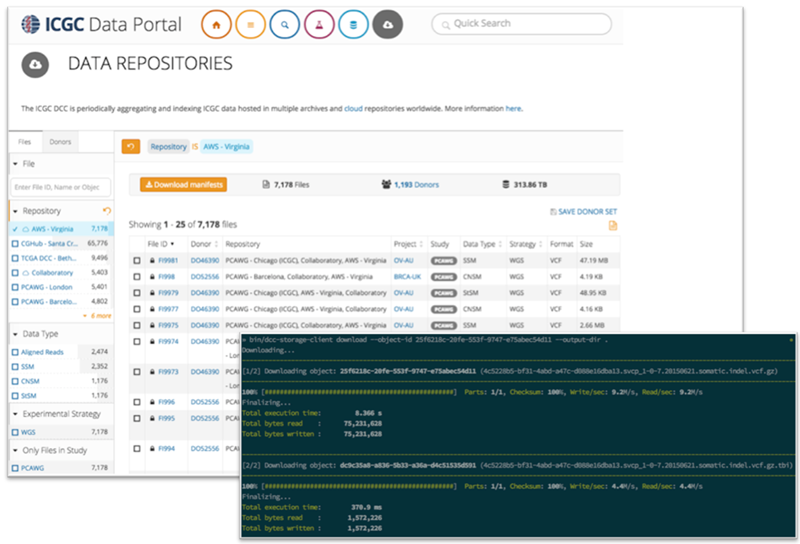
 The ICGC data will be generally accessible via the use of a downloadable command line tool. Users can search for files using the
The ICGC data will be generally accessible via the use of a downloadable command line tool. Users can search for files using the