My colleagues Jessica Beegle and Christopher Crosbie shared the inspiring story below!
-Jeff;
The science of cancer research is continually evolving to include new fields of study. Examples include development of chemotherapies, radiology amplified treatments, and epidemiology for identifying carcinogens. Pathology continues to help deepen the understanding of the disease's manifestations.
The discipline of computer science is a relatively new entrant in the quest to understand, treat and cure cancer. Computer science is needed to decipher how certain variations in our DNA relate to cancer and what treatment paths have the greatest potential for success for each individual. This type of task is best suited for computer science because the study of DNA, typically referred to as genomics, requires significant big data processing capabilities. In fact, scientists predict genomics will generate more digital information than astronomy, YouTube, and Twitter by 2025.
Today, much of the software developed to collect, analyze and visualize this data is created in silos among different IT systems, research departments, health care institutions, and even nations. This separation greatly hinders the speed of scientific discovery.
Researchers at Fred Hutchinson Cancer Research Center in Seattle wanted to change this. Led by Eric Holland, M.D., Ph.D., director of the Human Biology Division and Solid Tumor Translational Research at Fred Hutch, the team developed Oncoscape, an open-source web application to apply and develop analysis tools for molecular and clinical data. Oncoscape enables researchers to discover new patterns and relationships, which further cancer research.
To utilize current technology in computer science, the Oncoscape team collaborated with GitHub and AWS. The goal was to leverage the code-sharing platform that GitHub provides with the cloud computing capabilities that AWS offers. According to Dr. Holland:
 Hosting Oncoscape in the cloud makes it easy for our development team to make changes and redeploy the software in order to keep up with the needs of the research community. Knowing I can securely access the site from anywhere in the world allows me to show collaborators what is possible with data visualization and how we can use a common platform to work together in cancer research.
Hosting Oncoscape in the cloud makes it easy for our development team to make changes and redeploy the software in order to keep up with the needs of the research community. Knowing I can securely access the site from anywhere in the world allows me to show collaborators what is possible with data visualization and how we can use a common platform to work together in cancer research.
Robert McDermott, the IT Solutions Architect behind the AWS deployment of Oncoscape explains: “AWS is a very capable, reliable and flexible platform that allows us to quickly adapt to the needs of the project.” He cites maturity, reliability, breadth and depth of services and security as the key drivers for using AWS.
Oncoscape uses several AWS services including Amazon EC2, Elastic Load Balancing, Amazon CloudWatch, and Amazon S3. This approach makes it easy to distribute traffic across physical locations (Availability Zones), as well as quickly obtain actionable notifications in the event of a site issue. Amazon Route 53 has also proven useful for quickly making modifications to the development environment.
The diagram below depicts the full Oncoscape integration and deployment pipeline, including the merger points between GitHub, Circleci, DockerHub, Slack, and AWS.
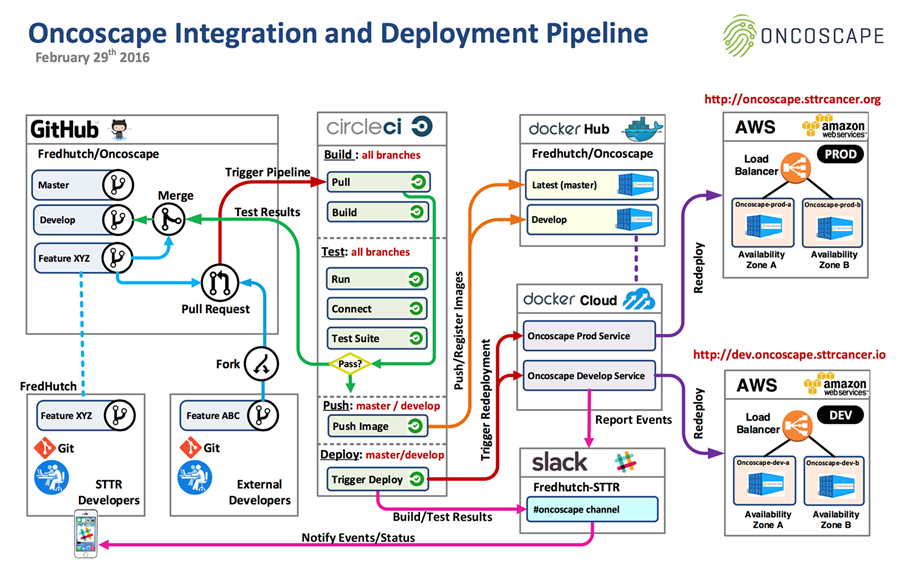
To learn more about the collaboration behind the Oncoscape project please watch this video or visit the Oncoscape home page.
- Jessica Beegle (Global Healthcare & Life Sciences Ecosystem Leader) and Christopher Crosbie (Healthcare and Life Science Solution Architect)



No comments:
Post a Comment